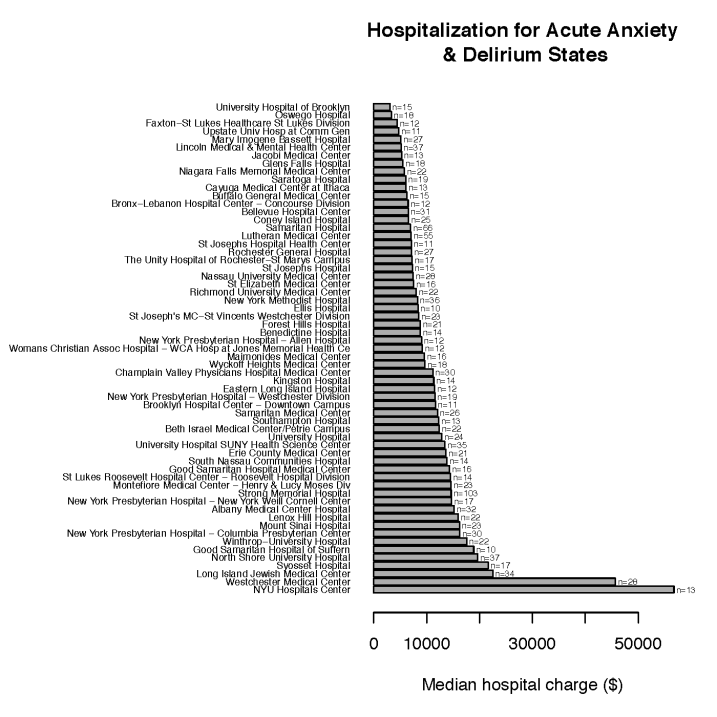
Many of us wonder how well health insurance payers cover genomic testing. The short answer is “not well” yet.
In 2014, the American Medical Association (AMA) issued new current procedural terminology (CPT) codes for genomic testing. These codes range from 81410 to 81471 (28 codes in total) and cover testing using targeted panel sequencing (5-50 genes), whole exome sequencing and whole genome sequencing (see Table 1). Codes for re-evaluation of previously obtained exome (CPT 81417) or genome (CPT 81417) sequence e.g, when updated knowledge is available or testing for unrelated condition/syndrome were even included. While the existence of such codes is a prerequisite for genomic testing reimbursement, health insurance payers do not automatically cover these tests.
Indeed the CMS fee schedule for these codes shows that only a very small fraction – 4 out of 28 – of these genomic testing procedures are associated with an actual reimbursement. This is only data from CMS but private payers typically follow the same fee schedule. The 4 covered procedures include targeted panel sequencing for oncology: CPT 81445 for solid tumor testing is reimbursed at $597.91 and CPT 81450 for hematological malignancies is reimbursed at $648.40. Surprisingly, panels capable of testing for both solid and heme malignancies (CPT 81455) are not reimbursed by CMS. Two complementary germline colon cancer risk assays covering ten genes or more (CPT 81435 for sequence analysis, CPT 81436 for duplication/deletion analysis) are reimbursed at $796.75.
These multi-gene genomic testing codes complement multi-gene expression assays with algorithmic analysis (0006M to 0010M, 81490 through 81595, 16 codes). Interestingly, 5 out of these 16 codes are reimbursed (Figure 1). This includes tests provided by Cologuard (CPT code 81528, $508.87) and Oncotype DX (CPT 81519, $3,419.42).
There are many CPT codes for testing of individual genes or pairs of genes and all such tests are reimbursed (Figure 1), with reimbursement levels ranging from $58.31 (PTEN gene, CPT 81322) to several thousand dollars. Interestingly a test simply covering Brca1&2 sequencing and full duplication/deletion analysis (CPT code 81162) is reimbursed at a whopping $2,485.86. Presumably this is linked to the ability of test providers to provide detailed interpretation of the BRCA1&2 test’s results, which in turn enhances clinical utility. Such interpretations may be facilitated by in-house proprietary databases of genomic variants such as the one that Myriad Genetics maintains. As clinical grade annotated genetic variant information becomes more broadly publicly available (see for example ClinVar, PCT, CivicDB, our own PMKB), it is likely that the clinical utility of multi-gene genomic testing will become more obvious.
Figure 1: CMS reimbursement for single and multi-gene assays.
Table 1: CPT codes for multi-gene genomic assays
| CPT code | Procedure description | 2016 CMS Fee ($) |
| 81410 | Aortic dysfunction or dilation (eg, Marfan syndrome, Loeys Dietz syndrome, Ehler Danlos syndrome type IV, arterial tortuosity syndrome); genomic sequence analysis panel, must include sequencing of at least 9 genes, including FBN1, TGFBR1, TGFBR2, COL3A1, MYH11, ACTA2, SLC2A10, SMAD3, and MYLK | 0 |
| 81411 | Aortic dysfunction or dilation (eg, Marfan syndrome, Loeys Dietz syndrome, Ehler Danlos syndrome type IV, arterial tortuosity syndrome); duplication/deletion analysis panel, must include analyses for TGFBR1, TGFBR2, MYH11, and COL3A1 | 0 |
| 81412 | Ashkenazi Jewish associated disorders (eg, Bloom syndrome, Canavan disease, cystic fibrosis, familial dysautonomia, Fanconi anemia group C, Gaucher disease, Tay-Sachs disease), genomic sequence analysis panel, must include sequencing of at least 9 genes, including ASPA, BLM, CFTR, FANCC, GBA, HEXA, IKBKAP, MCOLN1, and SMPD1 | 0 |
| 81415 | Exome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis | 0 |
| 81416 | Exome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis, each comparator exome (eg, parents, siblings) (List separately in addition to code for primary procedure) | 0 |
| 81417 | Exome (eg, unexplained constitutional or heritable disorder or syndrome); re-evaluation of previously obtained exome sequence (eg, updated knowledge or unrelated condition/syndrome) | 0 |
| 81420 | Fetal chromosomal aneuploidy (eg, trisomy 21, monosomy X) genomic sequence analysis panel, circulating cell-free fetal DNA in maternal blood, must include analysis of chromosomes 13, 18, and 21 | 0 |
| 81425 | Genome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis | 0 |
| 81426 | Genome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis, each comparator genome (eg, parents, siblings) (List separately in addition to code for primary procedure) | 0 |
| 81427 | Genome (eg, unexplained constitutional or heritable disorder or syndrome); re-evaluation of previously obtained genome sequence (eg, updated knowledge or unrelated condition/syndrome) | 0 |
| 81430 | Hearing loss (eg, nonsyndromic hearing loss, Usher syndrome, Pendred syndrome); genomic sequence analysis panel, must include sequencing of at least 60 genes, including CDH23, CLRN1, GJB2, GPR98, MTRNR1, MYO7A, MYO15A, PCDH15, OTOF, SLC26A4, TMC1, TMPRSS3, USH1C, USH1G, USH2A, and WFS1 | 0 |
| 81431 | Hearing loss (eg, nonsyndromic hearing loss, Usher syndrome, Pendred syndrome); duplication/deletion analysis panel, must include copy number analyses for STRC and DFNB1 deletions in GJB2 and GJB6 genes | 0 |
| 81432 | Hereditary breast cancer-related disorders (eg, hereditary breast cancer, hereditary ovarian cancer, hereditary endometrial cancer); genomic sequence analysis panel, must include sequencing of at least 14 genes, including ATM, BRCA1, BRCA2, BRIP1, CDH1, MLH1, MSH2, MSH6, NBN, PALB2, PTEN, RAD51C, STK11, and TP53 | 0 |
| 81433 | Hereditary breast cancer-related disorders (eg, hereditary breast cancer, hereditary ovarian cancer, hereditary endometrial cancer); duplication/deletion analysis panel, must include analyses for BRCA1, BRCA2, MLH1, MSH2, and STK11 | 0 |
| 81434 | Hereditary retinal disorders (eg, retinitis pigmentosa, Leber congenital amaurosis, cone-rod dystrophy), genomic sequence analysis panel, must include sequencing of at least 15 genes, including ABCA4, CNGA1, CRB1, EYS, PDE6A, PDE6B, PRPF31, PRPH2, RDH12, RHO, RP1, RP2, RPE65, RPGR, and USH2A | 0 |
| 81435 | Hereditary colon cancer disorders (eg, Lynch syndrome, PTEN hamartoma syndrome, Cowden syndrome, familial adenomatosis polyposis); genomic sequence analysis panel, must include sequencing of at least 10 genes, including APC, BMPR1A, CDH1, MLH1, MSH2, MSH6, MUTYH, PTEN, SMAD4, and STK11 | 796.75 |
| 81436 | Hereditary colon cancer disorders (eg, Lynch syndrome, PTEN hamartoma syndrome, Cowden syndrome, familial adenomatosis polyposis); duplication/deletion analysis panel, must include analysis of at least 5 genes, including MLH1, MSH2, EPCAM, SMAD4, and STK11 | 796.75 |
| 81437 | Hereditary neuroendocrine tumor disorders (eg, medullary thyroid carcinoma, parathyroid carcinoma, malignant pheochromocytoma or paraganglioma); genomic sequence analysis panel, must include sequencing of at least 6 genes, including MAX, SDHB, SDHC, SDHD, TMEM127, and VHL | 0 |
| 81438 | Hereditary neuroendocrine tumor disorders (eg, medullary thyroid carcinoma, parathyroid carcinoma, malignant pheochromocytoma or paraganglioma); duplication/deletion analysis panel, must include analyses for SDHB, SDHC, SDHD, and VHL | 0 |
| 81440 | Nuclear encoded mitochondrial genes (eg, neurologic or myopathic phenotypes), genomic sequence panel, must include analysis of at least 100 genes, including BCS1L, C10orf2, COQ2, COX10, DGUOK, MPV17, OPA1, PDSS2, POLG, POLG2, RRM2B, SCO1, SCO2, SLC25A4, SUCLA2, SUCLG1, TAZ, TK2, and TYMP | 0 |
| 81442 | Noonan spectrum disorders (eg, Noonan syndrome, cardio-facio-cutaneous syndrome, Costello syndrome, LEOPARD syndrome, Noonan-like syndrome), genomic sequence analysis panel, must include sequencing of at least 12 genes, including BRAF, CBL, HRAS, KRAS, MAP2K1, MAP2K2, NRAS, PTPN11, RAF1, RIT1, SHOC2, and SOS1 | 0 |
| 81445 | Targeted genomic sequence analysis panel, solid organ neoplasm, DNA analysis, and RNA analysis when performed, 5-50 genes (eg, ALK, BRAF, CDKN2A, EGFR, ERBB2, KIT, KRAS, NRAS, MET, PDGFRA, PDGFRB, PGR, PIK3CA, PTEN, RET), interrogation for sequence variants and copy number variants or rearrangements, if performed | 597.91 |
| 81450 | Targeted genomic sequence analysis panel, hematolymphoid neoplasm or disorder, DNA analysis, and RNA analysis when performed, 5-50 genes (eg, BRAF, CEBPA, DNMT3A, EZH2, FLT3, IDH1, IDH2, JAK2, KRAS, KIT, MLL, NRAS, NPM1, NOTCH1), interrogation for sequence variants, and copy number variants or rearrangements, or isoform expression or mRNA expression levels, if performed | 648.40 |
| 81455 | Targeted genomic sequence analysis panel, solid organ or hematolymphoid neoplasm, DNA analysis, and RNA analysis when performed, 51 or greater genes (eg, ALK, BRAF, CDKN2A, CEBPA, DNMT3A, EGFR, ERBB2, EZH2, FLT3, IDH1, IDH2, JAK2, KIT, KRAS, MLL, NPM1, NRAS, MET, NOTCH1, PDGFRA, PDGFRB, PGR, PIK3CA, PTEN, RET), interrogation for sequence variants and copy number variants or rearrangements, if performed | 0 |
| 81460 | Whole mitochondrial genome (eg, Leigh syndrome, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes [MELAS], myoclonic epilepsy with ragged-red fibers [MERFF], neuropathy, ataxia, and retinitis pigmentosa [NARP], Leber hereditary optic neuropathy [LHON]), genomic sequence, must include sequence analysis of entire mitochondrial genome with heteroplasmy detection | 0 |
| 81465 | Whole mitochondrial genome large deletion analysis panel (eg, Kearns-Sayre syndrome, chronic progressive external ophthalmoplegia), including heteroplasmy detection, if performed | 0 |
| 81470 | X-linked intellectual disability (XLID) (eg, syndromic and non-syndromic XLID); genomic sequence analysis panel, must include sequencing of at least 60 genes, including ARX, ATRX, CDKL5, FGD1, FMR1, HUWE1, IL1RAPL, KDM5C, L1CAM, MECP2, MED12, MID1, OCRL, RPS6KA3, and SLC16A2 | 0 |
| 81471 | X-linked intellectual disability (XLID) (eg, syndromic and non-syndromic XLID); duplication/deletion gene analysis, must include analysis of at least 60 genes, including ARX, ATRX, CDKL5, FGD1, FMR1, HUWE1, IL1RAPL, KDM5C, L1CAM, MECP2, MED12, MID1, OCRL, RPS6KA3, and SLC16A2 | 0 |
Note: CPT codes and descriptions are copyright 2016 American Medical Association. All rights reserved. CPT is a registered trademark of the American Medical Association (AMA).